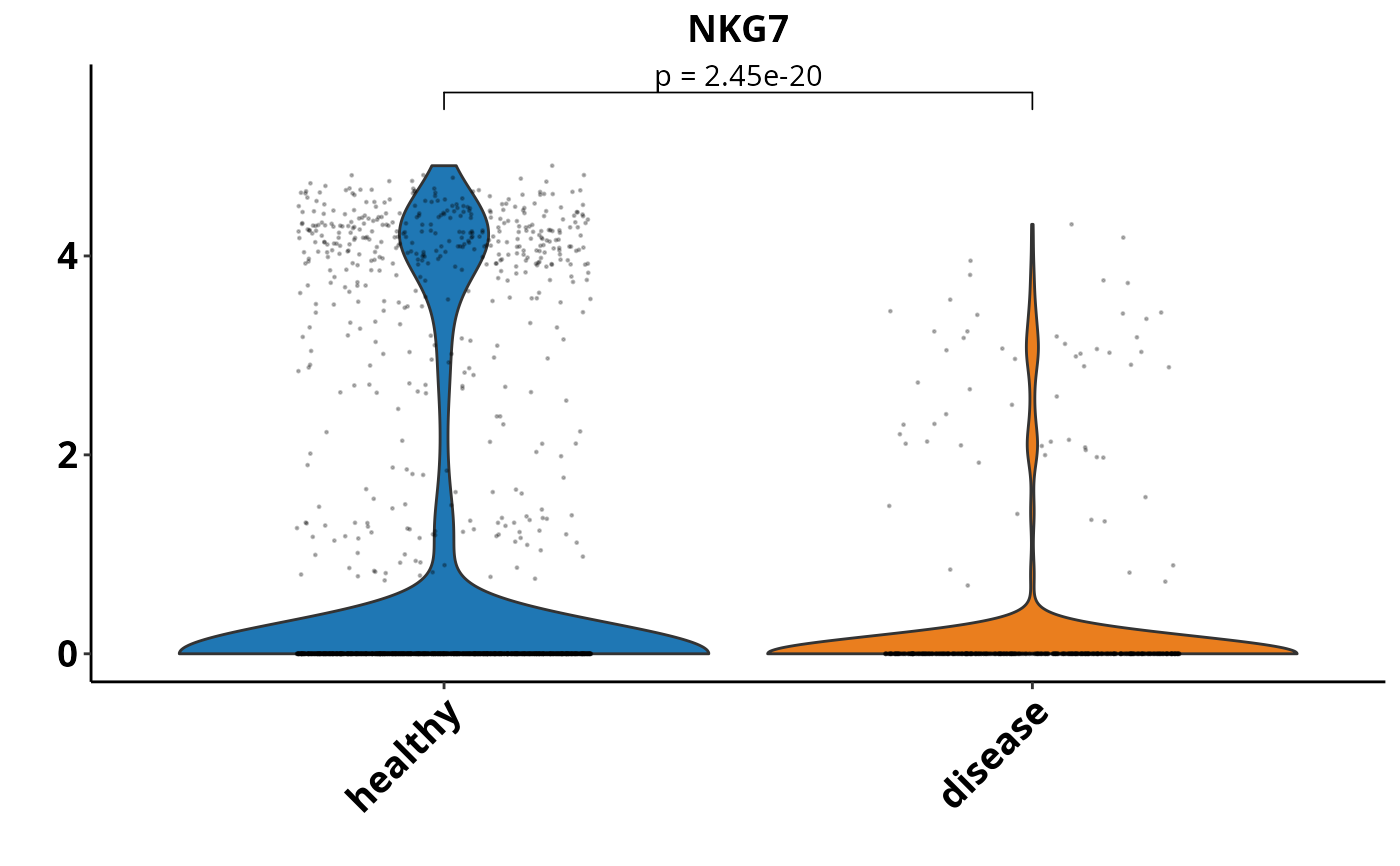
Creates a violin plot to compare gene expression across different conditions or groups within a Seurat object. It incorporates Wilcoxon rank-sum tests to evaluate statistical differences between conditions. The plot can be customized with options for data transformation, jitter display, and significance annotations. The function also supports multiple conditions and allows for visualisation of statistical results from wilcoxon test.
Usage
DO.VlnPlot(
sce_object,
SeuV5 = TRUE,
Feature,
ListTest = NULL,
returnValues = FALSE,
ctrl.condition = NULL,
group.by = "condition",
group.by.2 = NULL,
geom_jitter_args = c(0.2, 0.25, 0.25),
geom_jitter_args_group_by2 = c(0.1, 0.1, 1),
vector_colors = c("#1f77b4", "#ea7e1eff", "royalblue4", "tomato2", "darkgoldenrod",
"palegreen4", "maroon", "thistle3"),
wilcox_test = TRUE,
stat_pos_mod = 1.15,
hjust.wilcox = 0.8,
vjust.wilcox = 2,
size.wilcox = 3.33,
step_mod = 0,
hjust.wilcox.2 = 0.5,
vjust.wilcox.2 = 0,
sign_bar = 0.8
)Arguments
- sce_object
combined SCE object or Seurat
- SeuV5
Seuratv5 object? (TRUE or FALSE)
- Feature
name of the feature
- ListTest
List for which conditions wilcox will be performed, if NULL always CTRL group against everything
- returnValues
return df.melt.sum data frame containing means and SEM for the set group
- ctrl.condition
set your ctrl condition, relevant if running with empty comparison List
- group.by
select the seurat sce_object slot where your conditions can be found, default conditon
- group.by.2
relevant for multiple group testing, e.g. for each cell type the test between each of them in two conditions provided
- geom_jitter_args
vector for dots visualisation in vlnplot: size, width, alpha value
- geom_jitter_args_group_by2
controls the jittering of points if group.by.2 is specified
- vector_colors
specify a minimum number of colours as you have entries in your condition, default 2
- wilcox_test
Bolean if TRUE a bonferoni wilcoxon test will be carried out between ctrl.condition and the rest
- stat_pos_mod
value for modifiyng statistics height
- hjust.wilcox
value for adjusting height of the text
- vjust.wilcox
value for vertical of text
- size.wilcox
value for size of text of statistical test
- step_mod
value for defining the space between one test and the next one
- hjust.wilcox.2
value for adjusting height of the text, with group.by.2 specified
- vjust.wilcox.2
value for vertical of text, with group.by.2 specified
- sign_bar
adjusts the sign_bar with group.by.2 specified
Examples
sce_data <-
readRDS(system.file("extdata", "sce_data.rds", package = "DOtools"))
ListTest <- list()
ListTest[[1]] <- c("healthy", "disease")
DO.VlnPlot(
sce_object = sce_data,
SeuV5 = TRUE,
Feature = "NKG7",
ListTest = ListTest,
ctrl.condition = "healthy",
group.by = "condition"
)
#> SeuV5 TRUE, if object Seuratv4 or below change SeuV5 to FALSE
#> This message is displayed once per session.
#> Using group, orig.ident as id variables